Here’s the translation into American English:
—
A research team has made significant advancements in enzyme generation using generative artificial intelligence, specifically through protein language models (pLMs). This innovative strategy not only increases the natural diversity of these high-value biomolecules but also focuses on enhancing their stability, specificity, and effectiveness in human cells—crucial aspects for the advancement of biotechnology and medicine.
The company Metagenomi positions itself as a leader in the development of gene-editing therapies based on CRISPR enzymes. Its approach includes the use of an extensive database of natural enzymes, known as MGXdb, which enables them to identify promising candidates and train protein language models. These models can generate additional variants of certain classes of enzymes, which are later filtered through multi-model workflows to predict key characteristics and facilitate protein engineering campaigns.
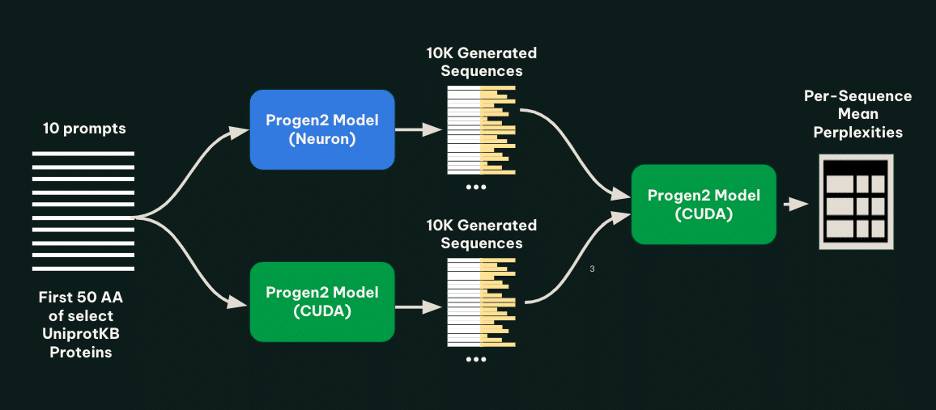
Recently, an article from the Metagenomi team revealed methods to reduce the costs associated with protein generation using the Progen2 model on AWS Inferentia instances. This approach has allowed for a reduction of up to 56% in the costs of generating variants compared to GPU-based technologies. Collaboration with Tennex and the AWS Neuron team has enabled the large-scale production of enzymes, a vital aspect of diversifying high-value enzyme classes.
The use of EC2 Inf2 instances has facilitated the large-scale execution of efficient inferences through AWS Batch, a service that optimizes the execution of hundreds of thousands of computational jobs efficiently. This infrastructure not only accelerates generation times but also minimizes expenses, a critical factor in research and development in the biotechnology field.
Comparative testing of different generation methods has shown that the modifications made do not compromise the accuracy of the models employed. These tests resulted in the generation of thousands of protein sequences, demonstrating that EC2 Inf2 Spot instances were significantly more cost-effective.
With the ultimate goal of generating millions of proteins, Metagenomi has leveraged its access to a vast metagenomic database and adapted its model to maximize both efficiency and effectiveness in enzyme generation. This effort not only marks an important advance in protein design, but also represents a milestone in cost reduction in biotechnology, potentially opening new opportunities for the production of high-value proteins in sectors such as pharmaceuticals and agriculture.
via: MiMub in Spanish