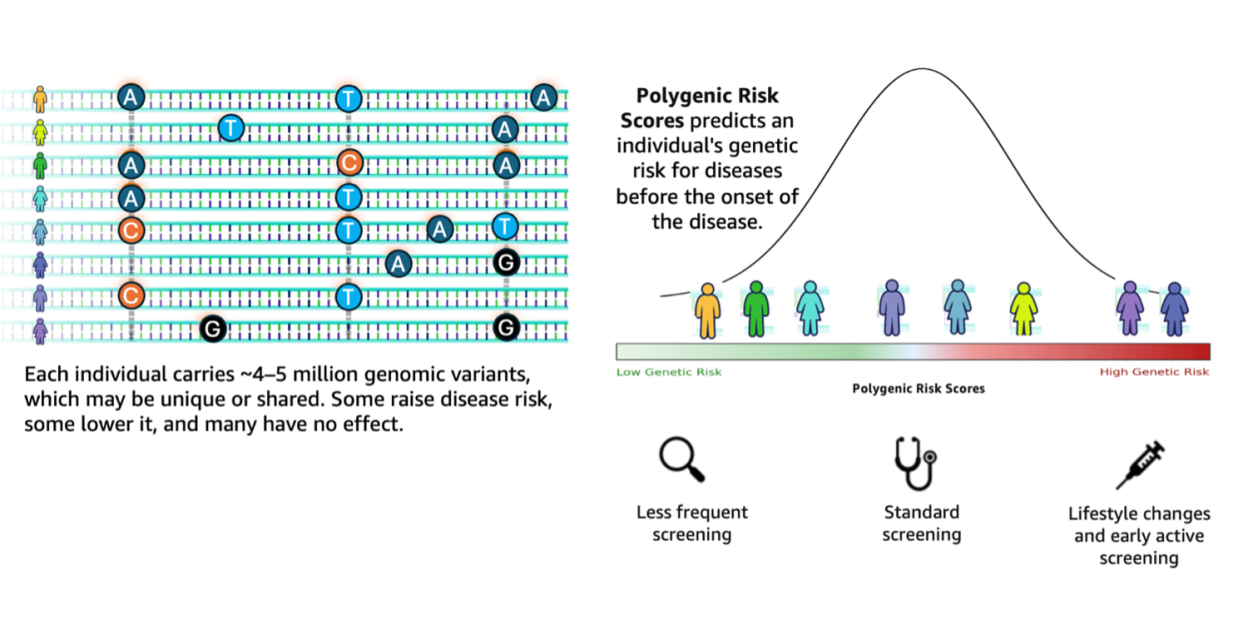
In the field of genetic research, there is a crucial transformation underway driven by the increasing amount of data generated through genome sequencing. A recent study from the 1000 Genomes Project indicates that a human genome has differences of between 4.1 and 5.0 million sites compared to the reference genome. These variants, predominantly single nucleotide polymorphisms (SNPs) and small insertions or deletions, have a significant impact on susceptibility to different diseases, a phenomenon that can be measured through polygenic risk scores (PRS). However, experts warn that workflows for analyzing this abundance of data face considerable challenges, making the process often fragmented and complex.
To address these obstacles, innovative solutions such as AWS HealthOmics are being implemented. This platform, along with tools like Amazon S3 tables and Amazon Bedrock AgentCore, is designed to facilitate the integration of variant files with understandable ontologies. Thanks to this, researchers can process data more efficiently, leading to more automated workflows. HealthOmics not only simplifies file format annotation (VCF), but also structures data in optimized formats, improving query performance on large variant cohorts.
One of the most notable advancements of these solutions is the use of a genomic variant interpretation agent, which combines intelligent analysis with automated processing. This agent allows researchers to ask questions previously reserved for technical experts, such as identifying patients with pathogenic variants in the BRCA1 gene or detecting drug resistance variations in specific cohorts. This removes a barrier that has limited genetic analysis to highly specialized bioinformaticians, empowering clinical researchers.
Furthermore, the system uses artificial intelligence to provide answers in a conversational format, making natural interaction with genetic data easier. Transforming raw variants into structured datasets, along with the ability to query in natural language, promises to improve access to critical information for clinical decision-making.
As technology advances, a future is envisioned where these applications integrate even more databases and external APIs, providing multimodal analysis that connects genetic data with clinical records. This evolution has the potential to not only enhance research, but also transform the approach to personalized treatments in healthcare.
The development of tools like these brings us closer to a more accessible and efficient future in obtaining valuable information about human genetics, significantly contributing to efforts in precision medicine and the progress of scientific discovery.
via: MiMub in Spanish